Despite all the attention genetics gets, it’s not DNA per se that controls the biological function of our body — it’s proteins.
The tiny, twisty molecules are to biology what atoms are to physics; they underlie all of the material and processes by which biological systems work, including the antibodies that play such a crucial role in the immune system.
Now, using a computer program called Rosetta, a team of researchers has developed a method for designing our own miniproteins.
“It’s pretty amazing stuff,” CalTech chemist Steve Mayo, who was not involved in the study, told Science’s Robert F. Service.
The miniproteins could lead to cost-effective and stable drugs for everything from cancer to COVID-19, and new diagnostic tests, as well.
Antibody issues: Antibodies are proteins designed to glom on to specific targets — e.g. gumming up SARS-CoV-2’s spike protein and hamstringing the virus’s attempt to enter your cells.
This makes them attractive and effective treatment options for a number of diseases, from infections to tumors.
While lab-made antibodies can be effective treatments for diseases like cancer and COVID-19, they have some drawbacks.
Antibodies are large proteins, which can make them expensive to create and potentially unstable, Science reported. You don’t want your treatment to fall apart before it can bind to its target.
Teams are learning how to create smaller, cheaper, and more stable antibodies, but there’s always been a tradeoff. Designing antibodies using only the target’s 3D structure “remains an outstanding challenge,” the researchers wrote in their study, published in Nature.
Getting into shape: Proteins noodle about like a ‘90s screensaver, spiraling and folding like a gymnast’s ribbon. And it is the shape of the proteins that is key, not just the amino acids they’re made of; get the shape right, and the body runs as normal. Get the shape wrong, like in prion diseases, and a protein could chew holes in your brain.
Ever since the American biochemist Christian Anfinsen floated the idea that shape makes the protein during his 1972 Nobel acceptance speech, researchers have tried to decode the protein’s structure — and predict how they will fold up based on their DNA sequence (a challenge just now cracked by deep learning AI).
The shape of proteins determines how other proteins — like antibodies, drugs, or diagnostic tags — will stick to them, and that determines how well they’ll work.
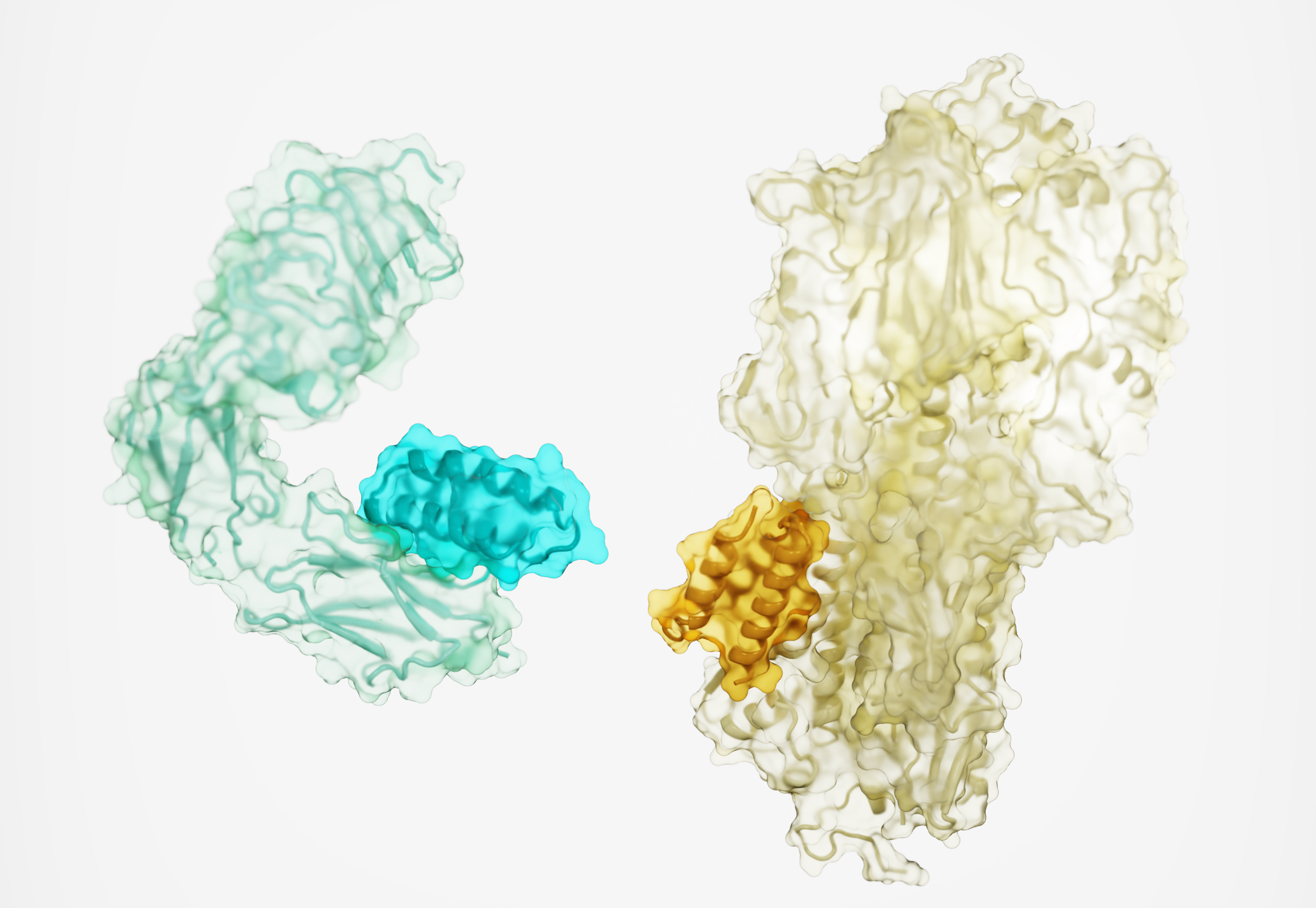
To enable miniproteins to stick onto targets as effectively as full-size antibodies, getting the proper shape would be crucial.
Think of it like a climbing wall, University of Washington computational structural biologist and corresponding study author David Baker told Science. There’s a smooth surface with a smattering of handholds and footholds, some stronger and easier to grasp than others. With their larger size, a normal antibody could snag a bunch of weak handholds and keep a good grip.
But because the miniproteins are smaller, the team decided that they needed to aim for strong handholds only — “hot spots” on the proteins that could give them just as much grip.
Problem is, for the vast majority of proteins, we don’t know where those hotspots are.
The Rosetta code: To identify potential hot spots for their miniproteins, the team turned to a software program called Rosetta, which was created to predict how a protein will fold based on its code.
The team first asked Rosetta to figure out how tightly certain amino acids will bind to various spots across a target protein, Science’s Robert Service explains.
That done, Rosetta then went about locating groupings of handholds — mapping out hot spots. Armed with that knowledge, Rosetta laid out the plans for a miniprotein.
Baker and colleagues created tens of thousands of digital versions of these miniproteins, Service reported, and tested them against simulated targets.
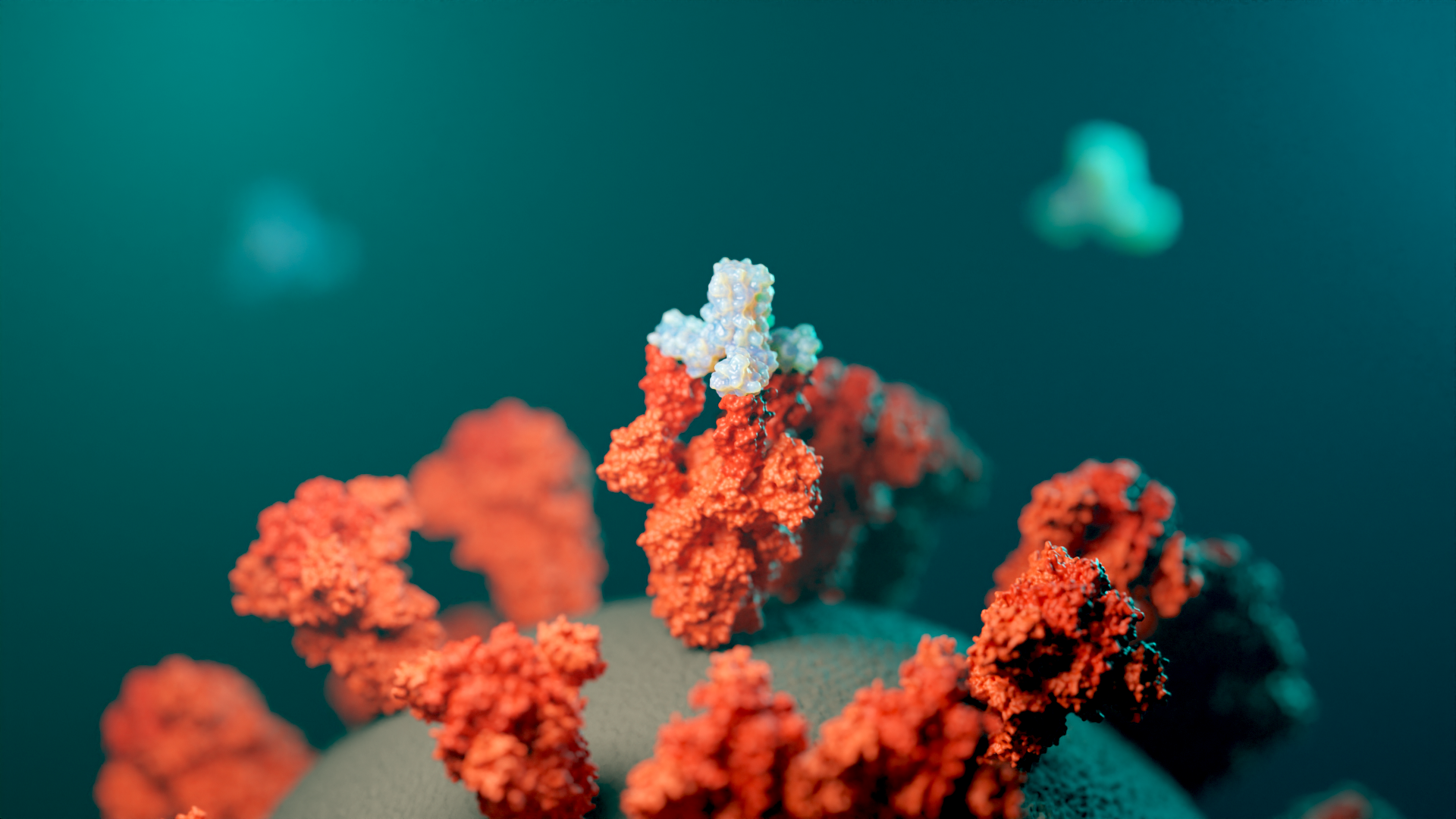
They then tested the miniprotein designs against 12 real targets, including proteins associated with cancer, SARS-CoV-2, and influenza. Not only did the miniproteins stick, but, with some tweaking, they were able to stick to the hot spots just as firmly as a larger antibody could — and sometimes stronger.
Don’t pop the champagne for miniproteins just yet, however; even with Rosetta lending a hand, many miniproteins didn’t stick at all. Trial and error is still a huge part of drug development, even with the incredible advances in AI and 3D modeling.
But as the technology improves, Baker expects the miniproteins to, as well.
“We can design proteins to bind to any target,” UW postdoc and co-author Longxing Cao told Service. The miniproteins could potentially block important virus proteins or carry cancer-killing payloads. They could also be used as a diagnostic tool, Tanja Kortemme, a UCSF bioengineer unaffiliated with the study, told Service.
“It opens up many possibilities.”
We’d love to hear from you! If you have a comment about this article or if you have a tip for a future Freethink story, please email us at [email protected].