As the world fights back against the COVID-2019 outbreak — now officially declared a pandemic by the WHO — the race is on for vaccines and antiviral medications that can help contain the virus.
A global challenge calls for global action, and the University of Washington’s Institute for Protein Design (IPD) is using a protein folding game, named Foldit, to bring everyone into the battle.
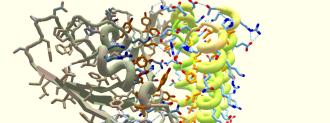
The game tasks players with solving puzzles by creating a protein structure that can interact with another protein in a given way. IDP is challenging their protein-folding gamers with something big: binding the coronavirus’s “spike protein,” which it uses to infect human cells — the first step in developing new antiviral drugs.
“Proteins do everything in the body,” says Brian Koepnick, a research scientist at the IPD. Proteins are made of chains of amino acids, and specific proteins cause all sorts of action inside the body, everything from metabolizing food to flexing muscles.
“We see lots of different solutions to solving this problem. That’s Foldit’s main advantage; our strong suit.”
Bruno Kestemont
Coronaviruses are studded with more proteins than a Hot Topic belt. (The name “corona” actually comes from the halo of protein spikes ringing the outside of the virus.) These proteins match up with specific proteins on the cells the virus evolved to infect. When the virus detects one of these proteins on human cells, it binds to it, allowing the virus to slip inside (the New York Times has a nice diagram of this).
Once inside, the viruses wrest control of the cell to replicate themselves, before escaping with a different protein (sometimes destroying the cell).
Foldit’s puzzle challenges players to create a protein that can stick to the virus’s spike, before it attaches itself to a human cell. Such a protein would help researchers create antiviral medications that could prevent the virus from infecting human cells — or at least slow it down.
Antiviral medications are not vaccines, which train the immune system to recognize threats and destroy them on sight. Nor are they like bactericidal antibiotics, which actually seek out and destroy bacteria. Instead, antiviral drugs are designed to interfere with the virus’s replication cycle, says Ashley Brown, program director of the Antiviral Pharmacodynamics Laboratory at the University of Florida.
The goal isn’t to stop virus replication completely, Brown says. “It’s more along the lines of to slow viral replication down to allow the immune system to come in and do what it needs to do.”
Antiviral medications can target any part of the replication cycle. Some aim to prevent the virus coming in, some to prevent it escaping, some to ensure the cell won’t work in the way the virus needs it to.
“We’re just trying to throw a wrench into the normal way that coronavirus infects human cells,” Koepnick says.
The difficulty is that there is an absolutely mind-melting amount of possible amino acid combinations that can make a protein, Koepnick says (*check the bottom for how many possibilities there are for even a tiny protein!). This vast number of potential answers makes finding the exact protein structure difficult.
“It would be a dream to be the one (or the group) who contributed to save humanity.”
Bruno Kestemont
This is where you come in. IPD’s solution is to get as many people trying to figure out the best protein for potential antiviral drugs as possible. The best protein structures that players come up with are analyzed by IPD and put through their paces; the hope is that they can work as well in real life as they do in the game.
The majority of Foldit’s players have no experience with proteins or some sort of biochemistry degree, Kopenick says. And this lack of specific training is actually a strength.
“I started playing when I saw a paper in a scientific news magazine (La Recherche) in 2012 where they said that players of a video game found — in only one week — the structure of a protein that challenged scientists for about 10 years,” says Bruno Kestemont, a Belgian Foldit player whose answer won round one of the spike protein problems in the soloist category.
“We see very creative solutions,” Koepnick says. “We see lots of different solutions to solving this problem. That’s Foldit’s main advantage; our strong suit.”
The problem this time: that damned spike protein. This is an especially tricky spike protein to bind, Koepnick says. Protein structures can contain water loving and/or water hating amino acids. Amino acids that hate water love to stick to each other, and these sticky sites are the best way to bind up a protein.
But SARS-CoV-2019 (the official name for the virus that causes COVID-19) is tricky, because its spike protein is not particularly sticky. According to Koepnick, there’s some good news though: only certain parts of the spike need to be blocked. If a protein can block the right site, it could be the basis for antiviral medications.
Antiviral development faces some tall hurdles even after a promising protein has been identified, Brown says. Researchers must ensure the drug gets to where it needs to go, and the proteins are not binding to places they shouldn’t — like our own blood cells. They need to make sure the antiviral does not cause adverse side effects or potentially damage the human cells. And because viruses are so specialized, they need to design drugs just as targeted.
And of course, antiviral medications need to be funded (preferably not after an outbreak has already happened). It all adds up to an expensive and time-consuming process — and the virus has a big head start.
“It’s always exciting to work on such an urgent problem. When we work on such a puzzle, more good players come back.”
Bruno Kestemont
The challenge — and the chance of contributing, in some natal way, to coronavirus antiviral drugs — means something not only to Koepnick and the Foldit creators, but also the players. This isn’t the first time they’ve worked on real world issues. Previous puzzles have featured such nasties as Ebola and Alzheimer’s.
“Of course, it’s always exciting to work on such an urgent problem. When we work on such a puzzle, more good players come back,” says Bruno Kestemont.
“It would be a dream to be the one (or the group) who ‘contributed to save humanity.’ But we know that it’s a far way from solving such a puzzle to finding the drug … to cure the disease.”
For a modest protein made up of 100 amino acids, there are 1,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,000,0000 possible combinations!