Since the 1970s, modern antibiotic discovery has been experiencing a lull. Now the World Health Organization has declared the antimicrobial resistance crisis as one of the top 10 global public health threats.
When an infection is treated repeatedly, clinicians run the risk of bacteria becoming resistant to the antibiotics. But why would an infection return after proper antibiotic treatment? One well-documented possibility is that the bacteria are becoming metabolically inert, escaping detection of traditional antibiotics that only respond to metabolic activity. When the danger has passed, the bacteria return to life and the infection reappears.
“Resistance is happening more over time, and recurring infections are due to this dormancy,” says Jackie Valeri, a former MIT-Takeda Fellow (centered within the MIT Abdul Latif Jameel Clinic for Machine Learning in Health) who recently earned her PhD in biological engineering from the Collins Lab. Valeri is the first author of a new paper published in this month’s print issue of Cell Chemical Biology that demonstrates how machine learning could help screen compounds that are lethal to dormant bacteria.
Tales of bacterial “sleeper-like” resilience are hardly news to the scientific community — ancient bacterial strains dating back to 100 million years ago have been discovered in recent years alive in an energy-saving state on the seafloor of the Pacific Ocean.
MIT Jameel Clinic’s Life Sciences faculty lead James J. Collins, a Termeer Professor of Medical Engineering and Science in MIT’s Institute for Medical Engineering and Science and Department of Biological Engineering, recently made headlines for using AI to discover a new class of antibiotics, which is part of the group’s larger mission to use AI to dramatically expand the existing antibiotics available.
According to a paper published by The Lancet, in 2019, 1.27 million deaths could have been prevented had the infections been susceptible to drugs, and one of many challenges researchers are up against is finding antibiotics that are able to target metabolically dormant bacteria.
In this case, researchers in the Collins Lab employed AI to speed up the process of finding antibiotic properties in known drug compounds. With millions of molecules, the process can take years, but researchers were able to identify a compound called semapimod over a weekend, thanks to AI’s ability to perform high-throughput screening.
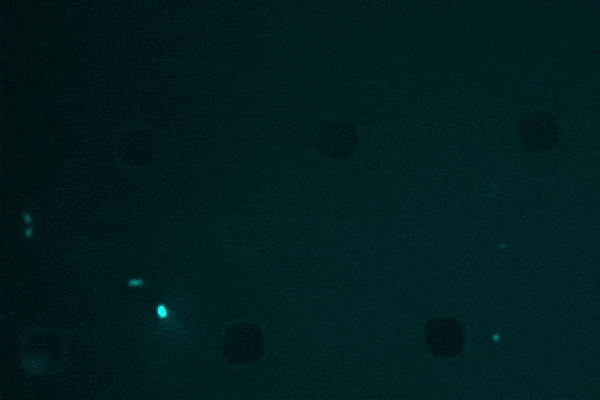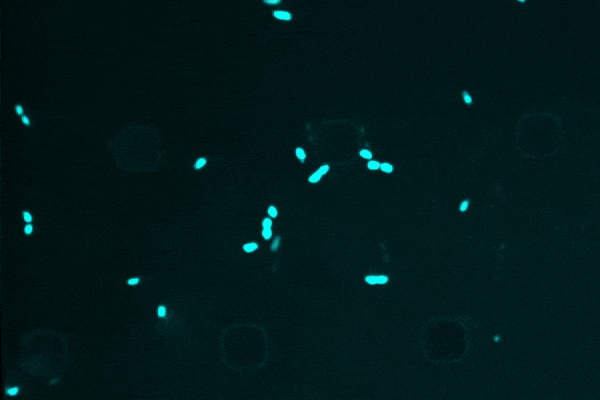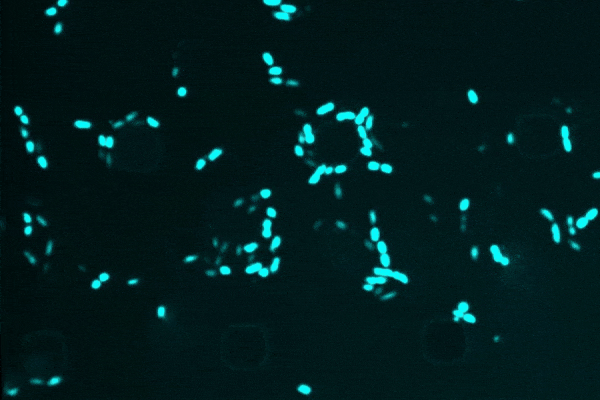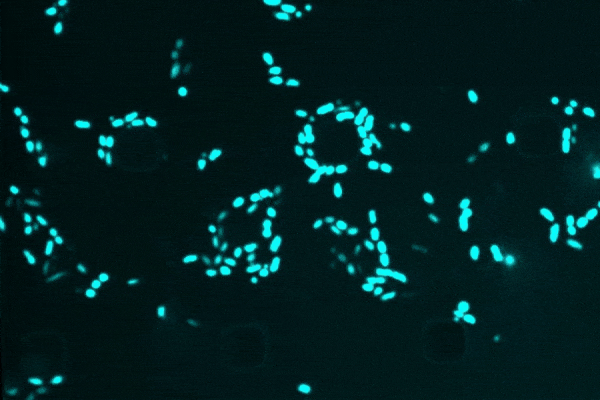
An anti-inflammatory drug typically used for Crohn’s disease, researchers discovered that semapimod was also effective against stationary-phase Escherichia coli and Acinetobacter baumannii.
Another revelation was semapimod’s ability to disrupt the membranes of so-called “Gram-negative” bacteria, which are known for their high intrinsic resistance to antibiotics due to their thicker, less-penetrable outer membrane.
Examples of Gram-negative bacteria include E. coli, A. baumannii, Salmonella, and Pseudomonis, all of which are challenging to find new antibiotics for.
“One of the ways we figured out the mechanism of sema [sic] was that its structure was really big, and it reminded us of other things that target the outer membrane,” Valeri explains. “When you start working with a lot of small molecules … to our eyes, it’s a pretty unique structure.”
By disrupting a component of the outer membrane, semapimod sensitizes Gram-negative bacteria to drugs that are typically only active against Gram-positive bacteria.
Valeri recalls a quote from a 2013 paper published in Trends Biotechnology: “For Gram-positive infections, we need better drugs, but for Gram-negative infections we need any drugs.”
Republished with permission of MIT News. Read the original article.





